I’m software developer with a special focus on scientific software.
In the past I joined multidisciplinar team focused on medicine and pathophysiology, modeling and simulation, high performance computation in grid and cloud computing networks. With my colleagues we have developed solution using backend technologies (.NET, ServiceStack, Java and Spring, recently Python, Flask) and frontend or backend-less technologies (HTML5, Javascript and standards WebAssembly, WebGL and libraries and frameworks Aurelia,babylonjs, Emscripten ).
I have a pleasure to work with experts in these scientific domains: computational physiology, integrative structural biology, psychoacoustic and auditory perception research.
Current projects
- eGolem - Virtual patient mannequin with realistic breathing, connected to ECMO device mockup and simulator of blood gases to train healthcare staff and show pathophysiology behind. egolem.online
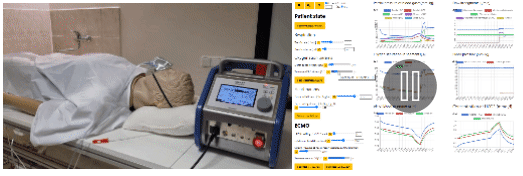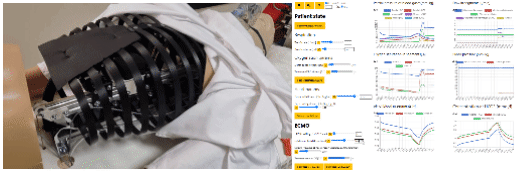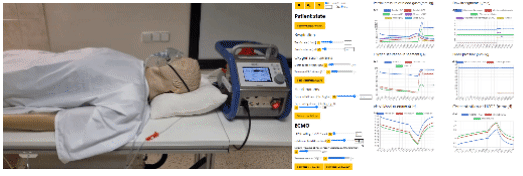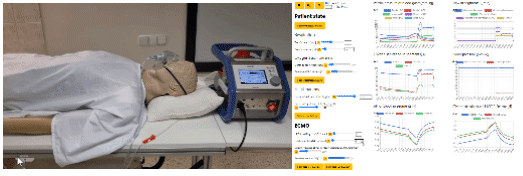
- Bodylight.js technology to transfer simple or complex mathematical model (Modelica) to scientific web simulator with modern web visualisation (WebAssembly, WebGL, Canvas) https://bodylight.physiome.cz

Past projects 2019-2022
- Virtual patient simulator, Robotic and mechanotronic trainer - supported by Czech ministry of industry and trade, led by COM SYS TRADE and partnered by Charles University, Creative Connections, Moravian Instruments, Inomech.
- Bodylight-docs - Tutorial and reference guide to Bodylight.js 2.0 beta (Modelica, WebAssembly, Web Components), open source tools to get fast model simulation and visualisation to browser.
- Bodylight-Scenarios - Bodylight powered scenarios of pathological physiology with simulation of models.
- Bodylight-VirtualBody - WEBGL powered 3D visualisation of human anatomy enriched with simulation of models of pathological physiology.
- Physiome.cz - basic research and education in computational physiology.
- Open source components - https://github.com/creative-connections/
Past projects
- B2NOTE - web tool for scientific data annotation led by e-Science Data Factory (SME) under EOSC-Hub project. Part WP 6.4 of EOSC-Hub - H2020 project led by EGI and partnered with many academic and industrial partners (developed B2Note version 2)
- H2020 West-Life - Virtual Research Environment for Structural Biology, European Horizon 2020 led by STFC UK in 2015-2018, data management work package and related parts, participated in WP6
- PROV-N-editor - ACE editor integrated with custom worker generated from ANTLR4 grammar with syntax highlighting and syntax validation. The editor can communicate with other web apps using cross document messaging API and can be filled with content using hash parameters ‘content’, ‘contentBase64’ and ‘contentLz’. Live - browser only demo
- sources of github.com/h2020-westlife-eu/virtualfolder - virtual folder view of scattered data, aggregates multiple data storage resource and provides unified way to access them, includes scripts to prepare VM template with basic structural biology software. (Frontend: HTML + Aurelia JS, backend: ServiceStack.NET)
- sources of github.com/h2020-westlife-eu/wp6-repository - exemplar implementation of repository of data for small and middle facilities in order to follow data management during life cycle of data in structural biology research. (Frontend: HTML + Aurelia JS, Backend: Java Spring)
- sources of github.com/h2020-westlife-eu/wp6-vm - Vagrant scripts to prepare various configuration of above components and tools.
- Virtual Patient - Physiovalues - web based system to perform simulation and parameter identification in multiple nodes distributed in local cluster or cloud computing environment
- Physiolibrary - Modelica library to enable and facilitate modeling of human physiology using mechanistic approach introduced in original HumMod and Physiomodel
- github.com/TomasKulhanek/Physiolibrary.models - various models of pulsatile and non pulsatile cardiovascular system in Modelica
-
Psychoacoustic Tests - web application to edit and perform assesment of perception on sounds, voice and music
- Medical Physiology in Modelica - 10% complete, aim to deliver free web based interactive book with models in modelica following common methods in modeling human physiology.
Short video made for crowdfunding campaign in 2015:
Talks and media
Pitch deck of eGolem, Wroclaw 2023, Poland - commercialization effort of a technology developed by our team
Talk at CERN (Geneva) in 2018 about use of CERN developed technologies in structural biology
about the project from http://cds.cern.ch/record/2302615
Short interview at EGI user forum (Manchester) in 2013
https://www.youtube.com/embed/hRlFrwQiN3o?si=Ger-BGo5YficmC_Y
Short interview at EGI user forum about relation of scientific domain (physiology) and technology (grid & cloud computing).
Academic work, articles in scientific journals or conference proceedings
- C.Morris, et al., West-Life: A Virtual Research Environment for structural biology, Journal of Structural Biology: X Volume 1, January–March 2019, 100006, https://doi.org/10.1016/j.yjsbx.2019.100006
- T. Kulhanek, C. Morris, M.D.Wynn (2017). West-Life Virtual Folder—Components and Tools. Instruct-Biennial 2017, http://doi.org/10.13140/RG.2.2.26963.78889
- T. Kulhanek, Dissertation
- T. Kulhanek, F. Jezek, M. Matejak, J. Silar, and J. Kofranek, ‘Experiences in teaching of m-odeling and simulation with emphasize on equation-based and acausal modeling techniques’, in 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 2015 [Online]. Available: http://dx.doi.org/10.1109/EMBC.2015.7319192
- F. Ježek, T. Kulhánek, K. Kalecký, and J. Kofránek, ‘Lumped models of the cardiovascular system of various complexity’, Biocybernetics and Biomedical Engineering, vol. 37, no. 4, pp. 666–678, 2017 [Online]. Available: http://dx.doi.org/10.1016/j.bbe.2017.08.001
- T. Kulhánek, J. Kofránek, and M. Mateják, ‘Modeling of short-term mechanism of arterial pressure control in the cardiovascular system: Object-oriented and acausal approach’, Computers in Biology and Medicine, vol. 54, pp. 137–144, Nov. 2014 [Online]. Available: http://dx.doi.org/10.1016/j.compbiomed.2014.08.025
- M. Mateják, T. Kulhánek, and S. Matoušek, ‘Adair-based hemoglobin equilibrium with oxygen, carbon dioxide and hydrogen ion activity’, Scandinavian Journal of Clinical and Laboratory Investigation, vol. 75, no. 2, pp. 113–120, Jan. 2015 [Online]. Available: http://dx.doi.org/10.3109/00365513.2014.984320
- M. Mateják, T. Kulhánek, J. Šilar, P. Privitzer, F. Ježek, and J. Kofránek, ‘Physiolibrary - Modelica library for Physiology’, in Proceedings of the 10th International Modelica Conference, March 10-12, 2014, Lund, Sweden, 2014 [Online]. Available: http://dx.doi.org/10.3384/ecp14096499
- T. Kulhanek, M. Matejak, J. Silar, and J. Kofranek, ‘Parameter estimation of complex mathematical models of human physiology using remote simulation distributed in scientific cloud’, in IEEE-EMBS International Conference on Biomedical and Health Informatics (BHI), 2014 [Online]. Available: http://dx.doi.org/10.1109/BHI.2014.6864463